HiCEnterprise is a software tool for identification of long-range chromatin contacts based on the Hi-C experiments. It was developed by Hania Kranas together with Irina Tuszyńska and Bartek Wilczyński. It implements three different statistical tests for identification of significant contacts at different scales as well as necessary functions for input, output and visualization of chromosome contact matrices. HiCEnterprise allow identifying chromosomal contacts on the level of enhancer-promoter interactions using Gumbell distribution (Won et al., 2016) as well as domain-to-domain interactions by hypergeometric (Niskanen et al., 2017), Poisson and negative-binomial distributions.
Citing HiCEnterprise
If you used HiCEnterprise in your research, please cite
Hania Kranas, Irina Tuszynska, Bartek Wilczynski, HiCEnterprise: Identifying long range chromosomal contacts in HiC data,
Code availability
The source code to the software is available on our github repository.
Minimum requirements
Python version 2.7+ or 3.5+, numpy, scipy, statsmodel, matplotlib libraries.
Installation
python setup.py install
Example
We provided some example files (used for testing too) with which you can learn to use the program. Here you can see two example runs:
Regions
In order to find all available options use
> HiCEnterprise regions -h
We used two Hi-C maps of chromosome 22 of Fetal brain cells: cortical plates (FBD) and germinal zone (FBP) from (Won et al., 2016), considering them replicates, we extracted them into separate numpy matrices that are available in the separate directories FBD and FBP.
After downloading, untar and unzip the mtx22.tar.gz file:
> tar -xvzf mtx22.tar.gz
Now two folders named FBD and FBP with Hi-C maps inside have appeared.
You can run the entire HiC map extraction procedure yourself :
First, get the data.
Then, untar and unzip the Fetal_maps.tar.gz file.
>tar -xvzf Fetal_maps.tar.gz
Next, extract numpy matrices for within-chromosome interactions, and rename the files to obtain files named “mtx-?-?.npy”, where “?” sign means chromosome number , e.g. with this python code and run:
>python extract.py
When HiC maps in the form of mtx-22-22.npy were already available, we have paired the Hi-C with enhancers from Fetal brain defined in Enhancer Atlas (Gao et al., 2016) to find regions interacting with those enhancers, and plotted results with matplotlib.
> HiCEnterprise regions –chr 22 –hic_folders FBD/ FBP/ –region_file ./Fetal_brain.fasta –bin_res 40000 –plotting mpl
Results are available to download here.
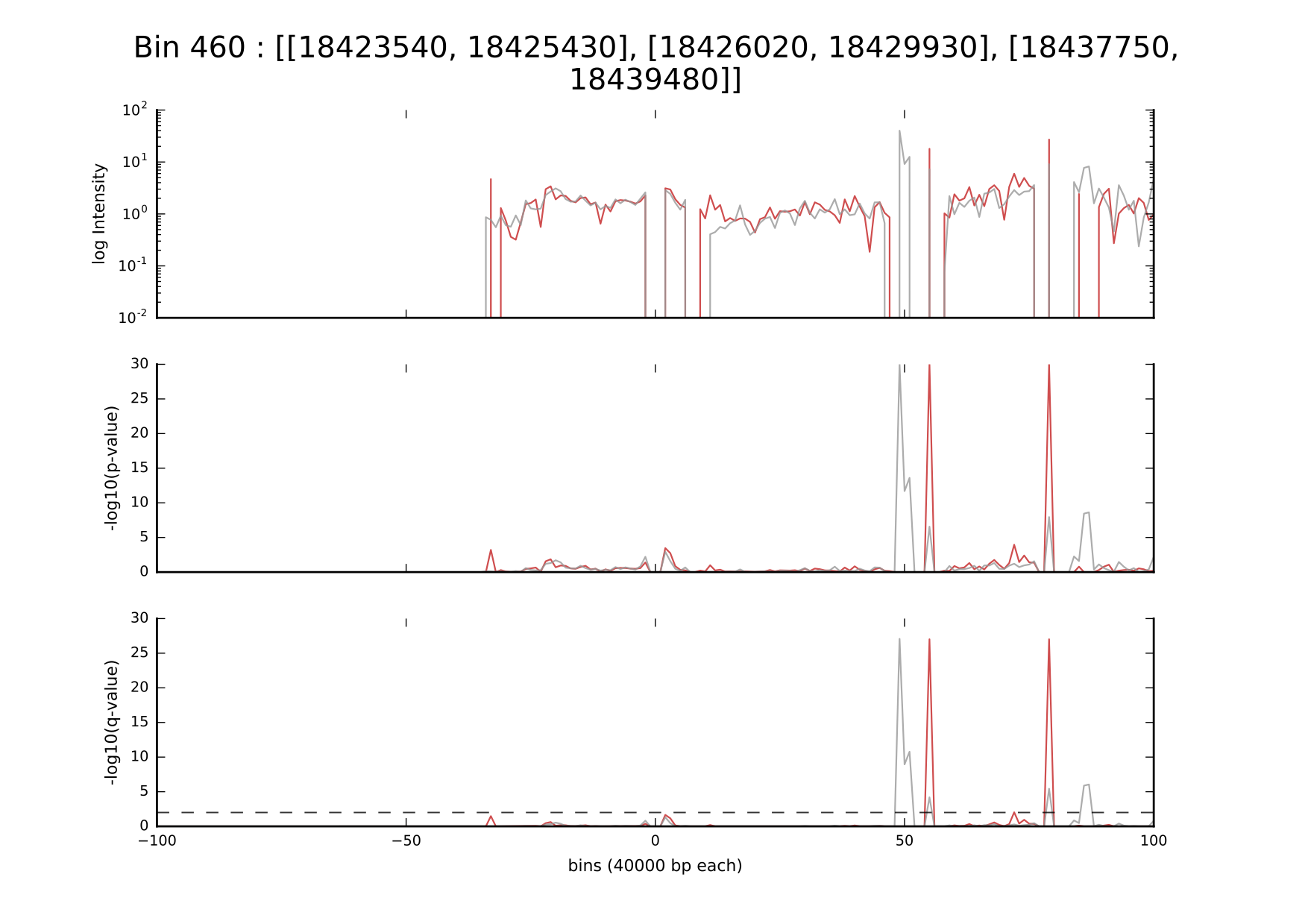
Below, you can see the plot for one of the chosen enhancers, showing the interactions, computed p-values, and corresponding q-values with the cutoff for significance.
Interestingly, 3 enhancers (18423540 – 18425430), (18426020 – 18429930), (18437750 – 18439480) located in bin 460 seem to interact significantly with 4 regions located further on the genome: two of them significant just in one of the cell types (grey), while two other ones are reproducible among both cell types, suggesting putative tissue specificity.
Domains
In order to find all available options use
> HiCEnterprise domains -h
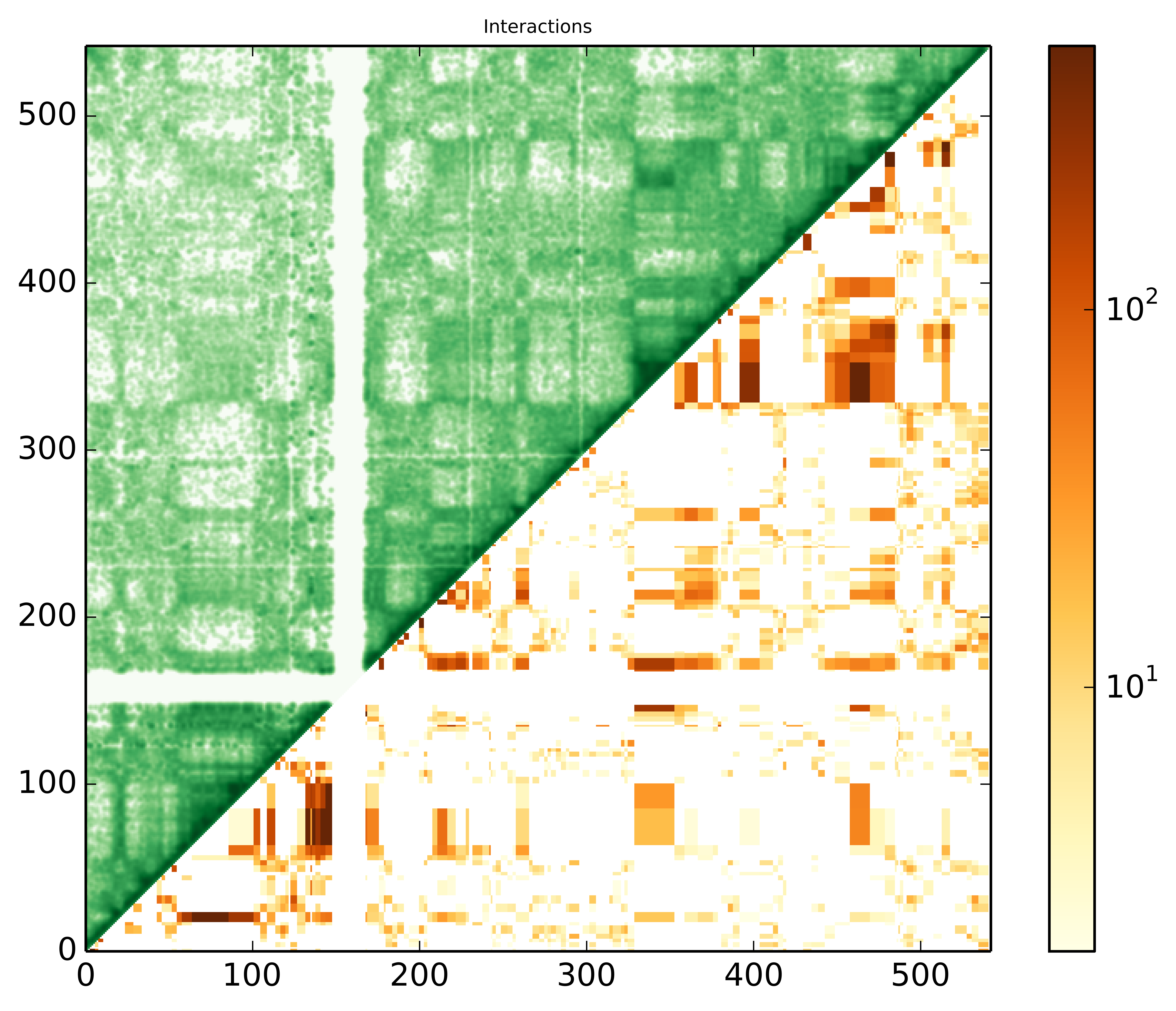
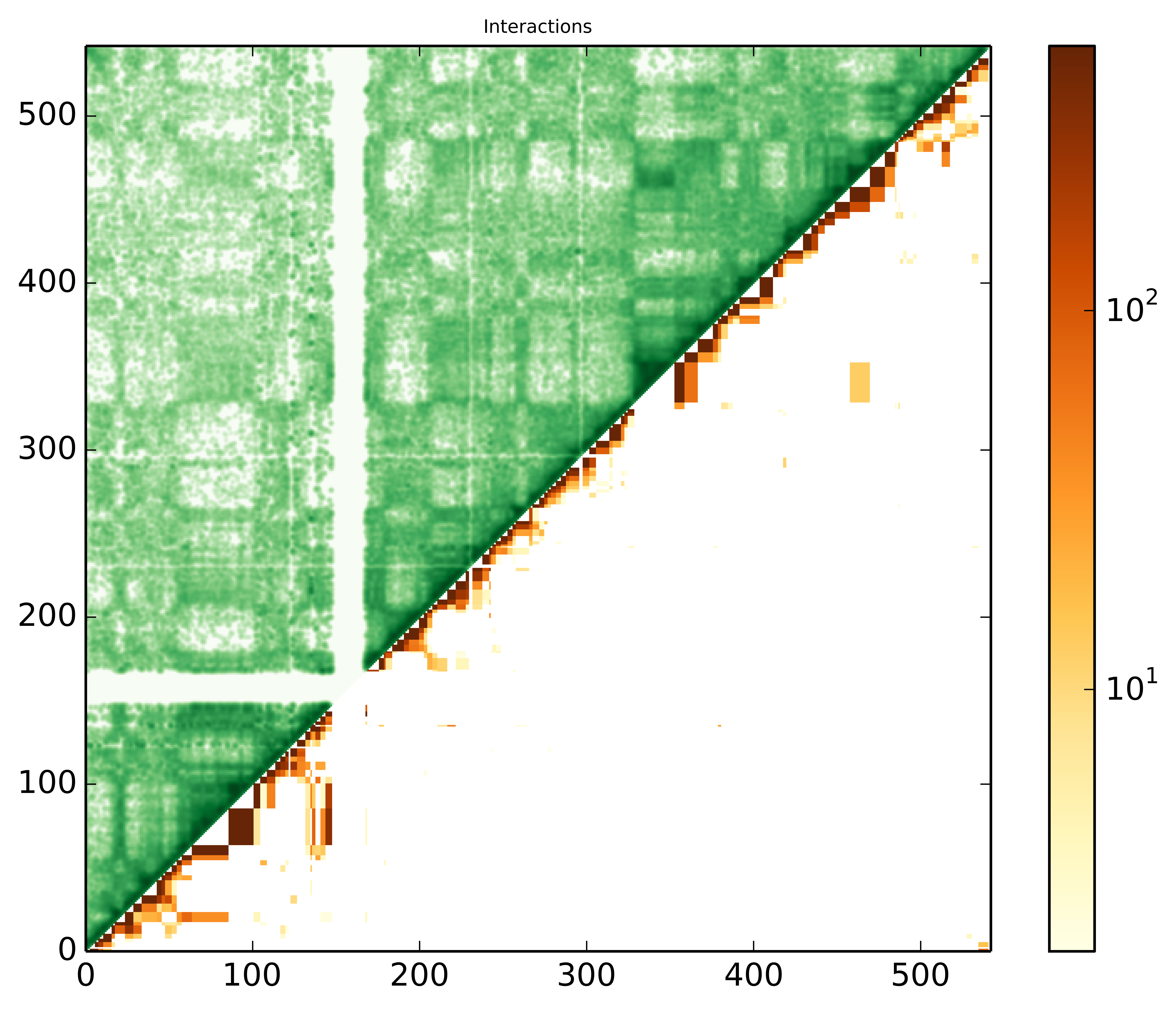
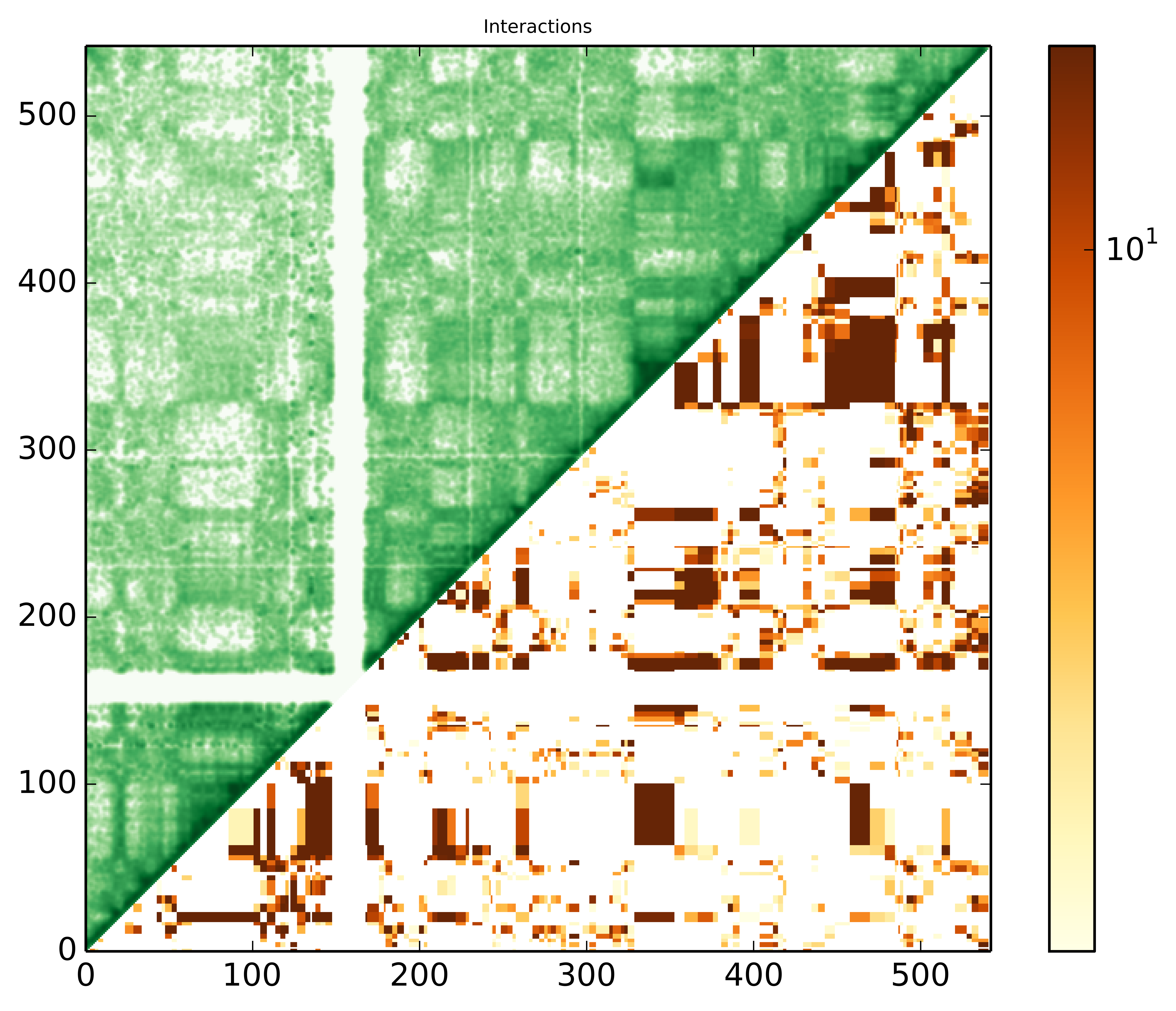
We used HiC map of 17th chromosome of human Endothelial cells (Niskanen et al., 2017) and domains defined by Sherpa softaware (https://github.com/regulomics/sherpa) to find domain-domain interactions:
> HiCEnterprise domains -c 1 -d HiCEnterprise/tests/test_files/doms/sherpa-tads –sherpa_lvl 2 -m HiCEnterprise/tests/test_files/maps/mtx-1-1.npy –plotting -b 150000
Due to default a distribution of HiCEnterprise is Poisson, we repeated calculations for hypergeometric and negative binomial distributions. Results are available here.
Domain-domain interaction for 17th chromosome of human Endothelial cells obtained using Poisson distribution
Domain-domain interaction for 17th chromosome of human Endothelial cells obtained using hypergeometric distribution
Domain-domain interaction for 17th chromosome of human Endothelial cells obtained using negative binomial distribution